Client
The RADx-rad Data Coordination Center Discovery & Diagnostics Core (RADx-rad DCC Dx Core), based at UC San Diego, is focused on closing the loop between data acquisition, structuring, analysis, data-driven modeling, decision support, implementation, evaluation, and dissemination at the point of care.
The National Institutes of Health (NIH) launched the Rapid Acceleration of Diagnostics (RADx®) initiative to speed innovation in the development, commercialization, and implementation of technologies for COVID-19 testing. By bringing together informaticians, data scientists and infectious diseases specialists to standardize viral samples, testing & procedures, and results, the goal was to integrate and share data in a meaningful manner.
Requirements & Context
One of the first critical missions of the scientific community during the COVID-19 pandemic was finding reliable methods for testing, and later finding ways of producing them at scale.
Manufacturers of these testing devices needed samples of the virus to conduct limit of detection (LOD) and challenge (distractor viral samples) tests, that could prove their devices could detect the virus in a variety of conditions before FDA approval. The RADx-radical program supported 49 teams across the USA.
The RADx-rad DCC Dx Core could provide these samples, and they needed a tool to create panels to create digital representations and hence obtain a centralized and homogeneous database that contained results from each different manufacturer, incoming from a variety of devices.
The RADx-rad DCC initiative was based on three pillars:
- Effective administration and coordination among programs;
- innovative approaches and tools to collect and standardize data and metadata to promote findability, accessibility, interoperability and reuse (FAIR) for data sharing; and
- principled preparation of standardized samples with known quantities of viral loads, and standardized procedures for testing new diagnostics to allow comparison across tests and calibration of new technologies.
Approach
Due to the urgency that a global scale pandemic implied, we partnered with InSTEDD to repurpose the Connected Diagnostics initiative, a project that used diagnostic data to improve healthcare delivery, outbreak detection and response, and health systems management.
Since the resulting product also served as a tool for labs and health workers, to provide better and faster treatment to diseases (faster notifications to patients that improve their treatment results), it was a great fit for the RADx-rad program.
Features
Labs need to cultivate each sample of a virus in batches, so the first task was to integrate a function in CDx to monitor batches. We also developed capabilities to print unique QR identifiers to label samples in the lab, giving personnel a way to quickly scan samples and access the information. Next we worked on a search engine for LOINC codes, to access specific information about each of the samples. Additionally, we built a tracking system that gave both UCSD and testing device manufacturers a transparent way of keeping track of these samples.
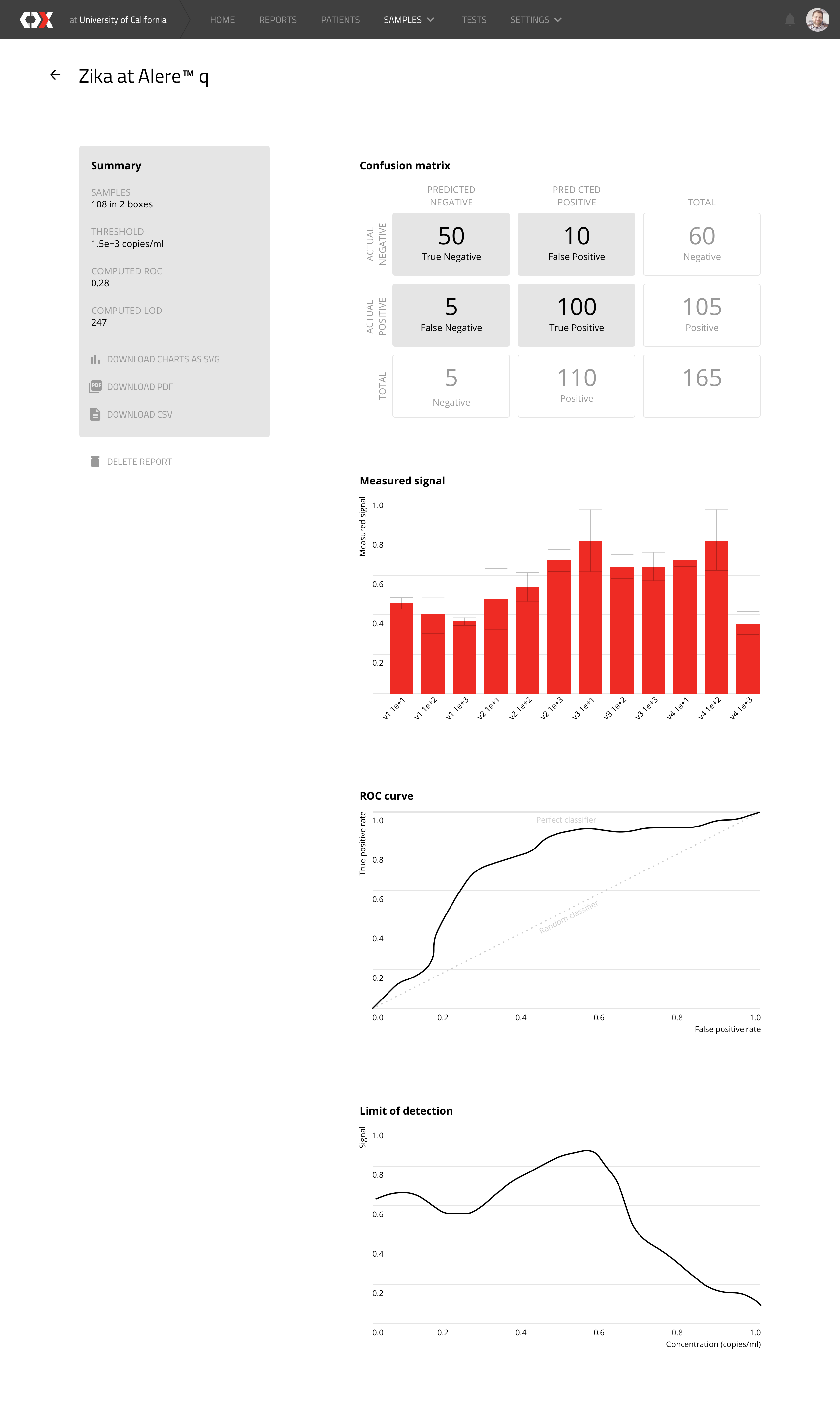
Another interesting feature is ‘blinding’: the system keeps information about each sample from the testing device manufacturers, so that they can process the data in an unbiased manner.Then, when all the obtained results are uploaded into the CDx platform, the samples are unblinded and the data is processed, analyzed and reported using automated analytical protocols. Based on the results obtained, the performance of the systems under development is assessed in terms of Limit of Detection, Sensitivity, and Specificity.
Results
We built an open source Laboratory Information Management System (LIMS) tool for creating panels of samples for limit of detection (LOD), challenge, and variant purposes. The end goal of the project was to have a live view of the virus’ evolution across various populations, keeping stats about location, etc. and to assess our ability to detect the variants using in-development Diagnostic Devices.
The RADx platform is capable of automatically generating test panels to be shipped out, then monitors the status of each sample (at the university, in transit, at the manufacturer). During 2023, we are now developing a feature to export visualization charts to files that labs can send to the FDA for approval of device homologation.
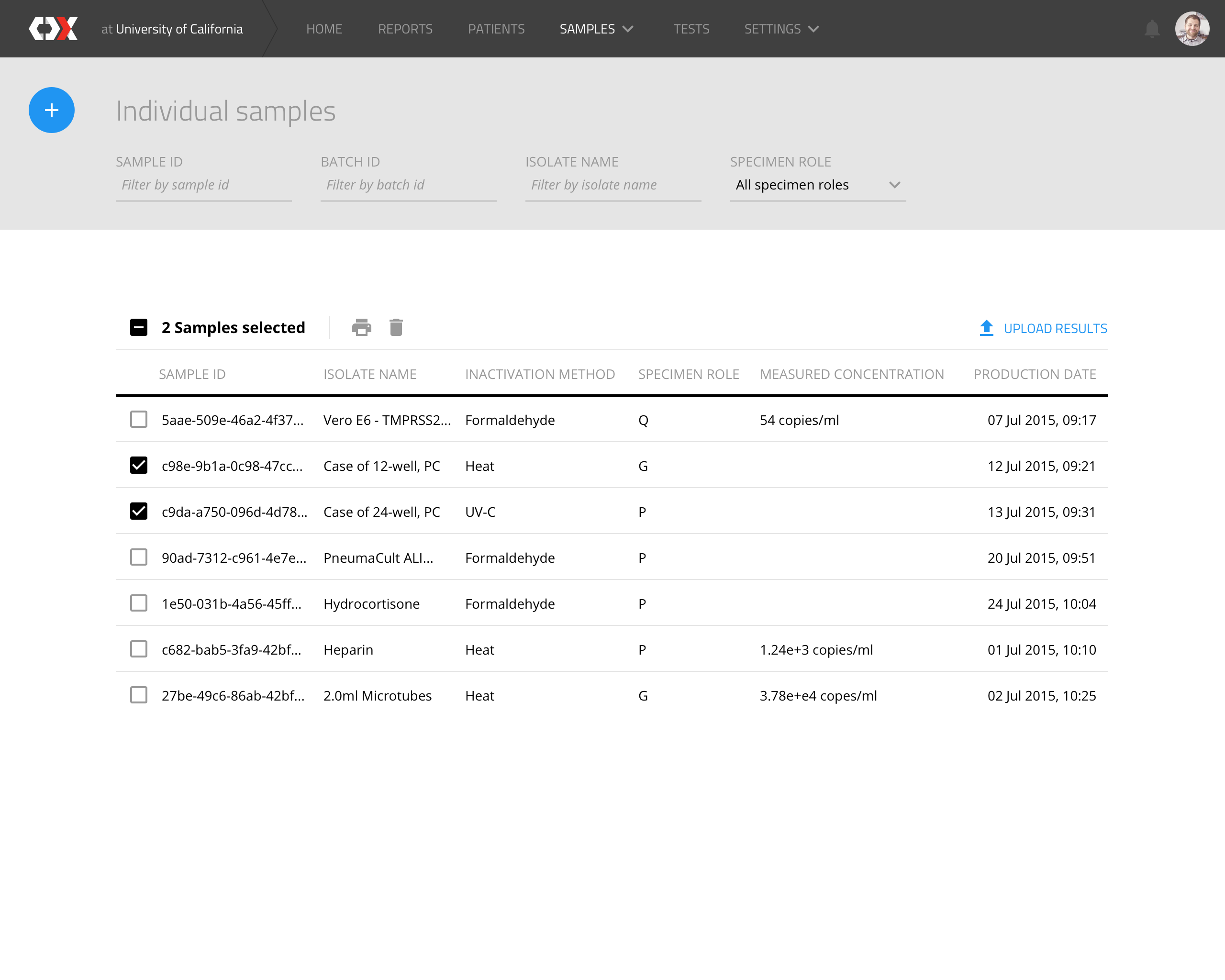
Individual samples
This view displays a list of all samples to be analyzed.
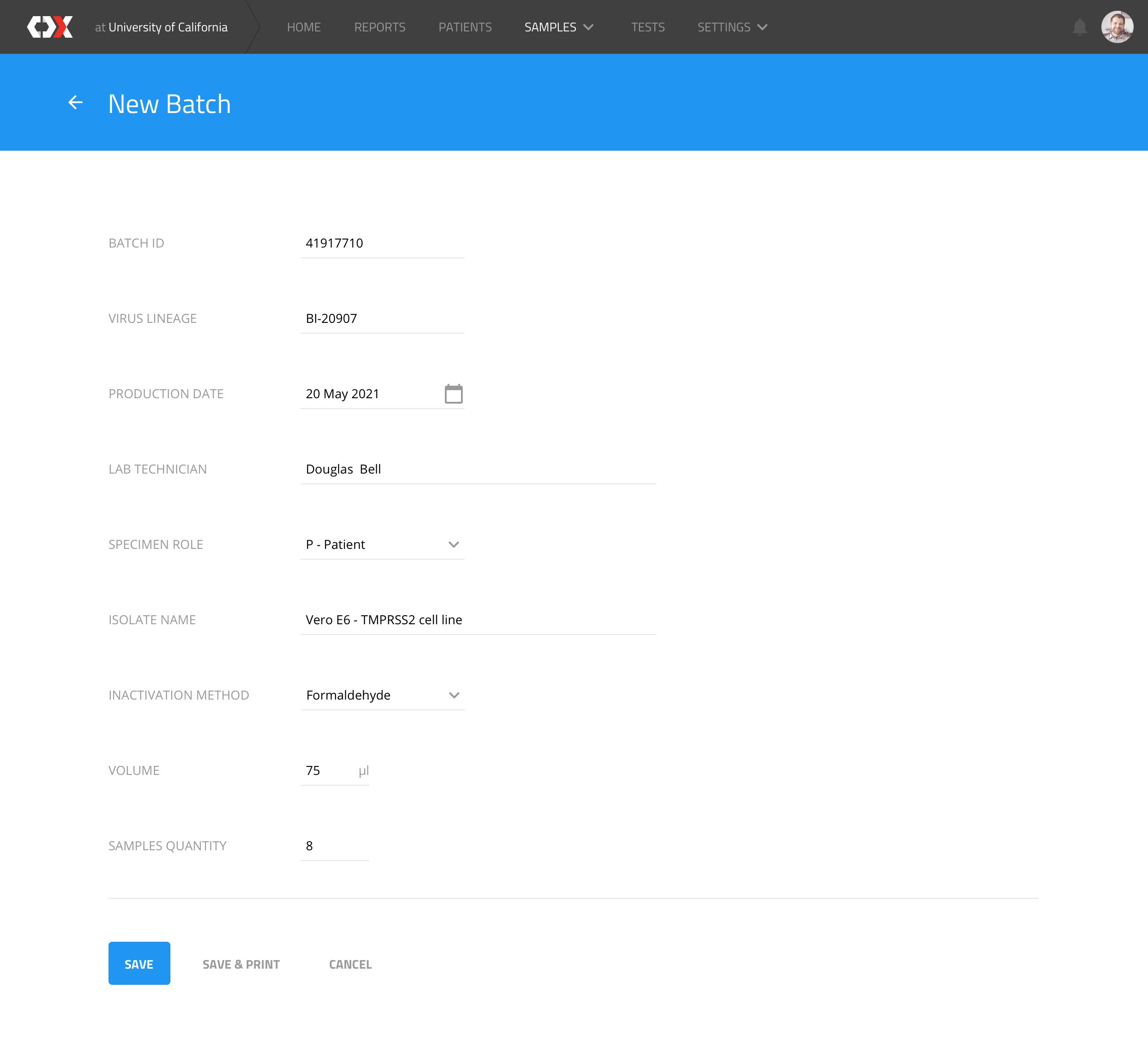
New batch
Users can create new batches to trigger the creation of N samples, according to trial requirements.
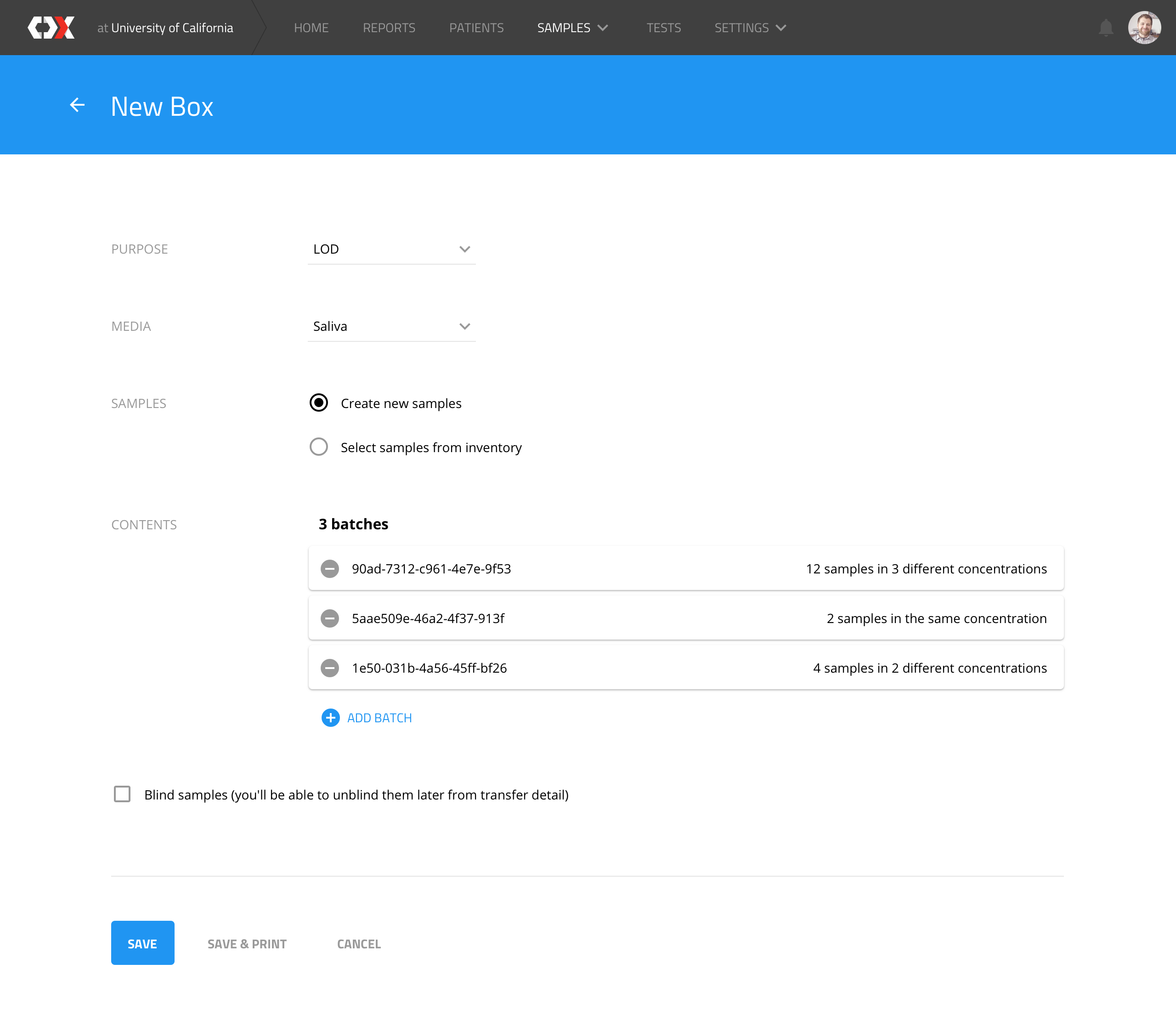
New box
Users can create an LOD (level of detection, i.e. the minimum virus concentration that a device can detect) box, which represents a physical box that is assembled with N samples from different batches.
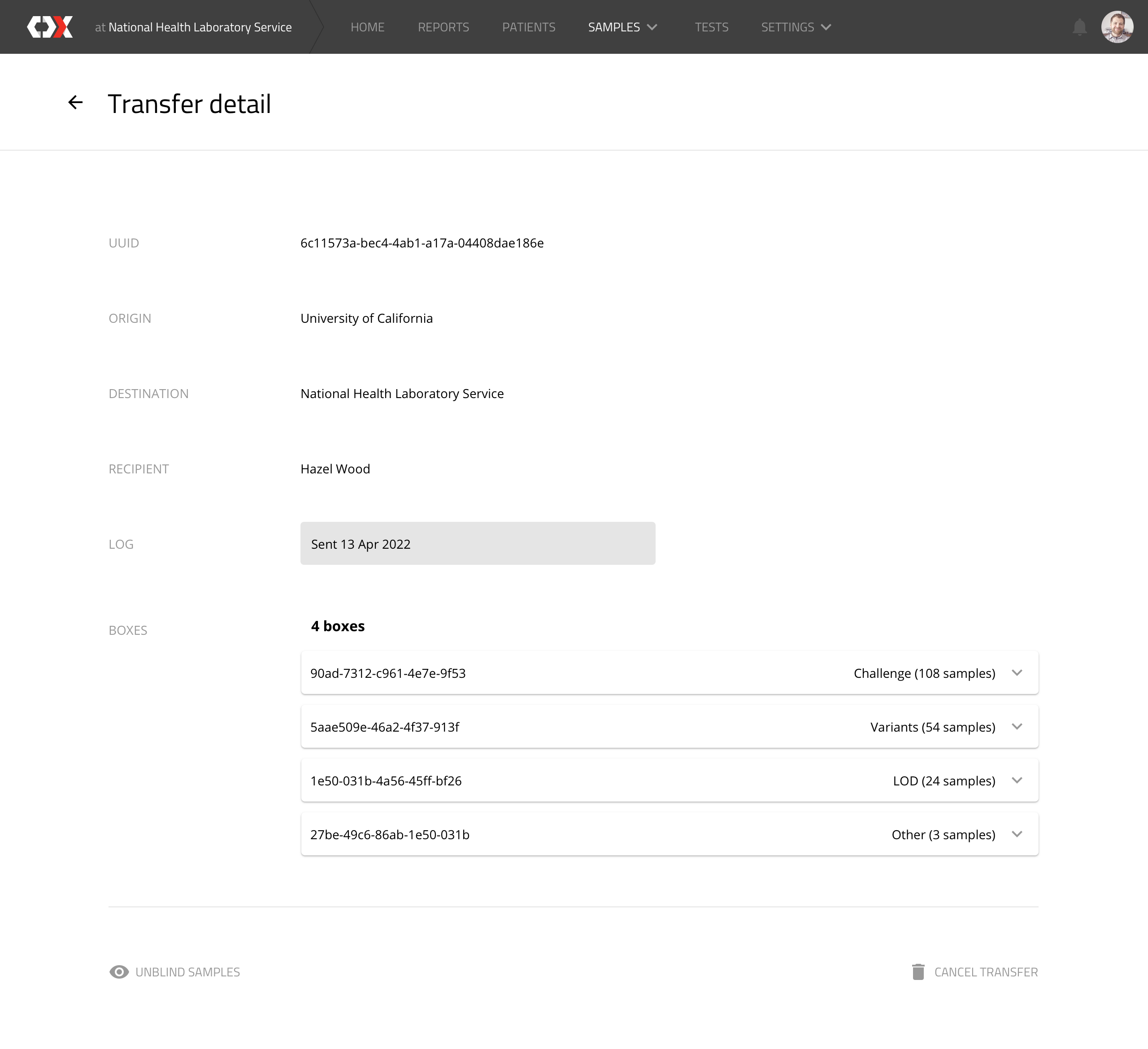
Transfer detail
This view displays transfer records. which represent the sending of a box to a device manufacturer.
Zika at Alere
This view shows how a device has performed in regards to the virus detection tests that was conducted on the box samples.
Open source
This project is Open Source, we invite you to collaborate and join us in the development of a better world through the use of technology.
https://github.com/instedd/cdxContributors

Nicolás di Tada Founder

Jonathan Kicillof Art Director

Matías García Isaía Full-stack Engineer & Site Reliability Engineer

Francisco Tarulla Alumni

Diego Liberman CEO

Diego Duarte Full-stack Engineer

Johannes Muller Principal Engineer

Leandro Radusky Alumni

Julien Portalier Principal Engineer

Mauricio Müller Alumni

German Batiston Alumni
Muhammad Usman Alumni

Carlos Stutz Zillner Alumni

Cecilia Lopez Alumni