Over the past few months, we embarked on a journey to bolster our expertise in the evolving domains of machine learning, deep learning, and bioinformatics. Our chosen method was to partake in the CAFA5 competition, where we grappled with predicting protein functions —an endeavor critical for comprehending the roles proteins play in diverse biological processes.
At its core, the competition centered on attributing probabilities to proteins, indicating their potential annotation with distinct functions represented by GeneOntology terms. In essence, the competition organizers furnished a collection of protein sequences, challenging participants to extrapolate new functions. Competitors could derive predictions solely from these sequential inputs or augment their models with any additional data sources.
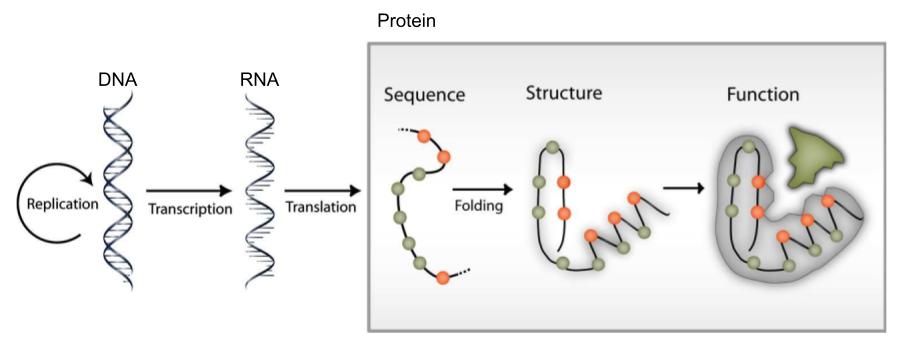
Information flow in molecular biology: DNA makes RNA, and RNA makes protein, which folds to form a protein structure, which determine protein function.
We developed a Python package facilitating the automated retrieval of varied biological data from standard databases. This encompassed protein sequences and metadata from UniProt, protein structures from PDB and AlphaFoldDB, and protein functions from GeneOntology. Subsequently, to manipulate this information, we delved into the rich Python bioinformatics ecosystem, leveraging, e.g., libraries like BioPython for encoding protein sequences and calculating structural contact maps, and obonet for managing graph-based relationships pertaining to GeneOntology protein functions.
With the data already processed and formatted for model feeding, we harnessed diverse neural network architectures to construct classifiers within our package. Ultimately, we honed our abilities to create classifiers for individual GO terms using:
- One-Dimensional Convolutional Networks (1D-CNNs) fueled by one-hot encoded protein sequences.
- Recurrent Neural Networks (RNNs) fed with protein sequences.
- Two-Dimensional Convolutional Networks (2D-CNNs) for constructing models based on protein structure contact maps.
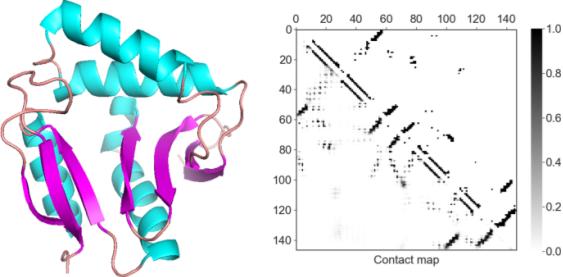
A protein structure visualized as a cartoon (left) and as a residue 2-D contact map(right).
While developing, we devoted considerable time to read and learn about the diverse strategies used in previous editions of the competition and the approaches adopted by other participants in the higly active discussion board of the competition.
Our objective was to prognosticate the likelihood of proteins executing specific functions, drawing from the training data supplied by the competition organizers. Each protein function carried a distinct score tied to the information accrued by it (more details here). Guided by this, we channeled our energies into designing models for terms with higher scores and iteratively refining them.
Through multiple iterations, we introduced a consensus score to enhance prediction robustness. This amalgamated scores from diverse models, yielding predictions for each protein-function pair. This proved enhancing as certain protein function models relied more heavily on structural insights compared to others (based on available data). By factoring in model precision and structural data reliability, our consensus score improved the final accuracy of our predictions.
The ultimate assessment of our performance hinged on how well our predictions aligned with experimentally ascertained protein functions amassed throughout the four-month competition period. Our final score settled at ~0.49, akin to the values attributed to baseline protein embeddings, a strategy chosen by the majority of the participants. While we read and learned about this strategy, we chose favoring the learning of more general deep learning approaches that could prove useful in a wider range of potential scenarios in our day-to-day work at Manas.
For those interested, our project repository can be found here. It contains tutorials that guide you through utilizing the tools we’ve developed: from manipulating biological data to training and evaluating neural networks accuracy. We hope that this work will serve as a useful resource for those interested in the intersection of machine learning and bioinformatics.
